With ALS-ENABLE, you have access to multiple structural biology methods. We can help you with each technique, and can help you combine techniques to maximize the information you can obtain about your system of interest.
Learn more about each technique below. Several of the methods are also described on the BER Structural Biology Portal.

Solution X-ray Scattering (SAXS)
Solution X-ray scattering, often called small-angle X-ray scattering (SAXS), provides structural information about any solution sample. Most investigators use SAXS to characterize the behavior of macromolecules in solution at the nanometer scale. It is an ideal assessment tool in the iterative design cycle of macromolecular engineering and can be used to characterize the degree of ordering in heterogeneous materials such as degrading cellulose.
Solution X-ray scattering (SAXS) enables high-throughput structural characterizations of gene products in solution.
SAXS can distinguish between aggregated and unfolded proteins, define global structural parameters and oligomeric states for most samples, identify shapes and structures similar to unknown structures, and determine protein envelopes.
READ ARTICLE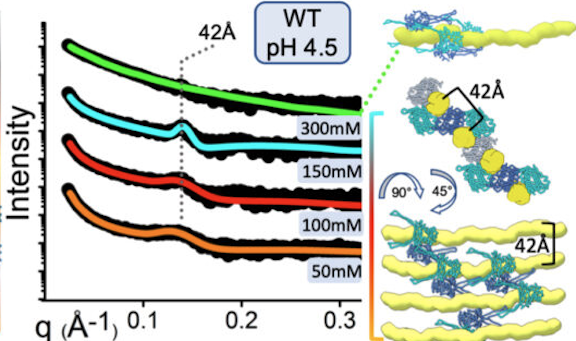
SAXS reveals how protein-mediated reorganization of microbial DNA adapts E. coli to a changing environment.
Experimental SAXS curves (left) indicate that HU, a histone-like nucleoid-associated protein (blue and cyan structures at right), organizes the structure of the nucleoid (yellow) differently at a high salt concentration (300mM) than at lower concentrations (50-150mM). This, in turn, affects global gene regulation in the cell.
READ ARTICLE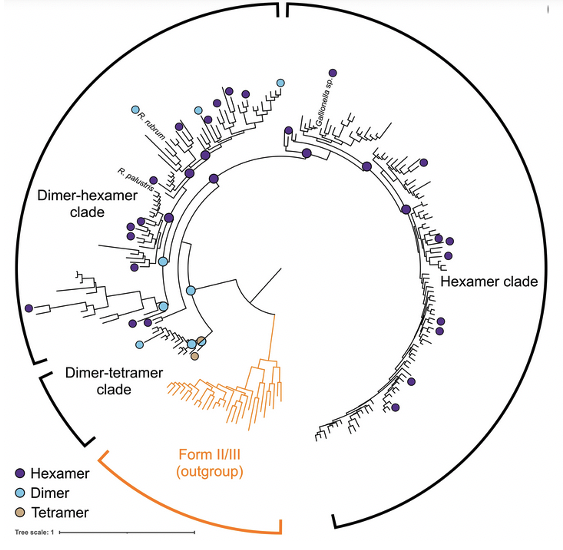
Diversity-driven sampling reveals plasticity of RuBisCo oligomeric state
By using SEC-SAXS-MALS researchers were able to conduct a diversity-driven structural study across extant and ancestral form II rubisco and retrace the evolutionary trajectory taken during the evolution of the enzyme.
READ ARTICLE
X-ray footprinting
Chemical footprinting has been used for over five decades to use solvent accessibility as a way to determine structural information on proteins and nucleic acids. The idea of using X-rays to for footprinting, however, is relatively new. X-ray footprinting was first conceived and tried out at the National Synchrotron Light Source at Brookhaven National Laboratory in the late 90’s. Since then, the technique has grown in popularity and been used to study a wide range of protein and nucleic acid systems, from small proteins to large complexes to ribosome motion within cells. The method is now available at the ALS beamline 3.3.1. We use X-rays to irradiate low protein concentration solutions; the X-rays interact with water molecules and create hydroxyl radicals, which covalently modify protein sidechains which are solvent accessbile. This “water map” is then used to infer protein structure. Contact us to learn more about using this method.
Macromolecular crystallography
Macromolecular crystallography (MX) is a widely-used standard method to obtain high resolution structural information on proteins and protein/nucleic acid assemblies. The ALS-ENABLE group runs 8 MX beamlines, with varying capabilities.
EXPERTISE
When you submit a proposal for MX, we will help you determine the best beamline and data collection strategies for your project.
COLLABORATIVE CRYSTALLOGRAPHY
We offer data collection services for users at many different levels, from collection through analysis.
AUTOMATION
The MX beamlines offer automounters, automatic loop centering, and a automated data collection and analysis. Contact us to find out more about puck types and accessing remote collection capabilities.
SPECIALIZED MX
Several of the MX beamlines are set up for room temperature measurements and plate-based measurements. Contact us to find out more about the range of capabilities and how to access them.